RuleSet Explanation

This document provides detailed instruction for preparing the files, and steps to run CypRules. CypRules is easy to use and only requires the specific structure file format.

CypRules Server predicts the metabolizing Cytochromes P450 (CYPs) inhibition, including CYP1A2, CYP2C19, CYP2C9, CYP2D6 and CYP3A4. CypRules predicts its results based on C5.0 algorithm. The rules are calculated based on Mold2 2D descriptors. The suggested ruleset that leads to the predicted outcome will be listed as well. This server will be helpful for researcher working in the field of drug discovery. We hope to contribute to decreasing the cost of discovering new drug molecules.

CypRules accepts only the SDF file format. SDF is one of a family of chemical-data file formats developed by MDL; it is intended especially for structural information. "SDF" stands for structure-data file, and SDF files actually wrap the molfile (MDL Molfile) format. Multiple compounds are delimited by lines consisting of four dollar signs ($$$$). A feature of the SDF format is its ability to include associated data. Here is an example SDF file containing five chemical structures.

After you get the SDF file ready, the CypRules will be right executing after you click the button "Upload". The execution time mainly depends on the size of the screening SDF file due to internet connection upload speed and the descriptor calculation speed. It's has been tested that a SDF containing 6000 chemical structures works well.

Here We'd like to provide a few SDF files for you to try out your own virtual screening!
A SDF file containing 100 chemical structures.
A SDF file containing 1000 chemical structures.
A SDF file containing 8 CYP1A2 known Inhibitors.

The result of CypRules contains mainly three information. The visualization of the compound structure, the prediction as inhibitor or non-inhibitior and the rulesets.
When CypRules finished the prediction process, a result page will be displayed as follows:
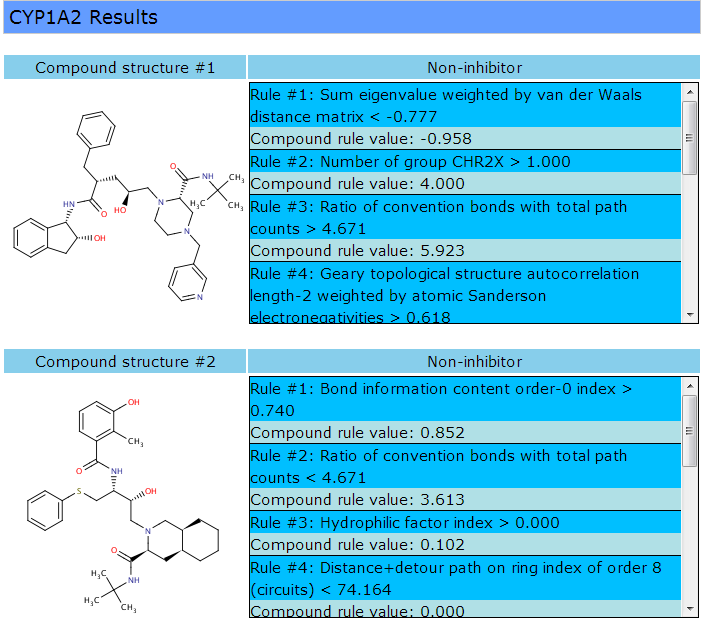

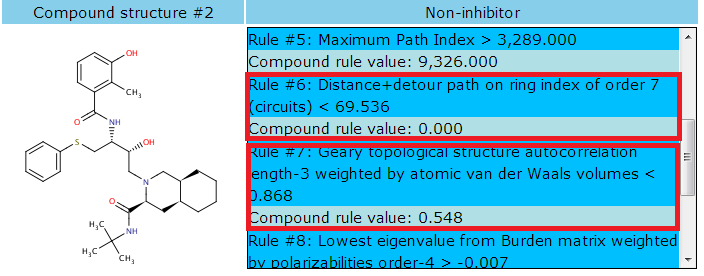
The prediction for the selected P450 endpoints is either inhibitor or non-inhibitor. As shown in the red square, calculated descriptors of the molecule qualify the certain inhibitor or non-inhibitor model rules.
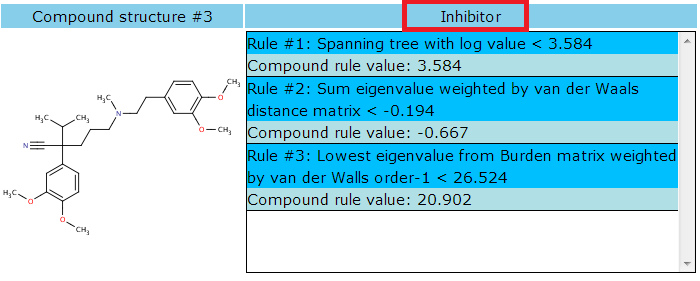
The rules that the given compound qualifies are listed sequentially with rule numbers. If the rules exceed the range of the area, a scroll bar will appear for further displaying. The row in deep sky blue shows the rule and the rule condition. The row in powder blue show the compound's actual calculated value of the rule.